Welcome to Xscape
What is Xscape?
Xscape is a set of Python tools for maximum parsimony phylogenetic event-based tree reconciliation in undated trees using the DTL (duplication-loss-transfer) model with applications to gene trees and species trees; parasite trees and host trees; and species trees and area cladograms.
Rather than using explicit event costs, these tools find solutions that are Pareto-optimal with respect to event counts; reconciliations are identified by event count vectors that count the number duplications, transfers, and losses that they incur. A reconciliation is Pareto-optimal if there is no other reconciliation with a strictly better event count vector.
Our algorithms efficiently compute the set of all Pareto-optimal solutions. Using these solutions, the event cost space is partitioned into regions. Each region has an associated event count vector and all event costs in that region admit the same set of maximum parsimony reconciliations (whose event counts are given by the region's event count vector). In other words, these algorithms relate event costs to the resulting reconcilations. The Xscape tool suite provides three tools that use these algorithms in novel ways.
The tools are based on results described in our paper:"Pareto-Optimal Phylogenetic Tree Reconciliation," R. Libeskind-Hadas, Y-C Wu, M. S. Bansal, and M. Kellis, Bioinformatics, Volume 30, Issue 12, June 15, 2014 (Proceedings of ISMB 2014)
Errata and Bug Fixes
Errata to the paper and bug fixes can be found on the errata page. The current version of the software is 0.0.5 (released August 23, 2015).What do the tools do?
All three tools assume that speciation is a "null" event of cost 0. (Although the underlying algorithms can be adapted for the general case that speciation has a non-zero cost and even negative cost.) Since the event costs are unit-less, duplication is normalized to cost 1. Transfer and loss are therefore the two free variables and are assumed to have positive cost. The cost landscape is therefore 2-dimensional space of the transfer and loss costs. All three tools take the following input (described in more detail in the README file accompanying the software):
- The name of a file containing the species (host) tree and the gene
(parasite) tree in newick format, followed by the associations
between their leaves (one line per association in the format
gene:species).
- The name of an output file where the results will be saved.
- A numerical range for the loss and transfer costs.
The three Xscape tools are:
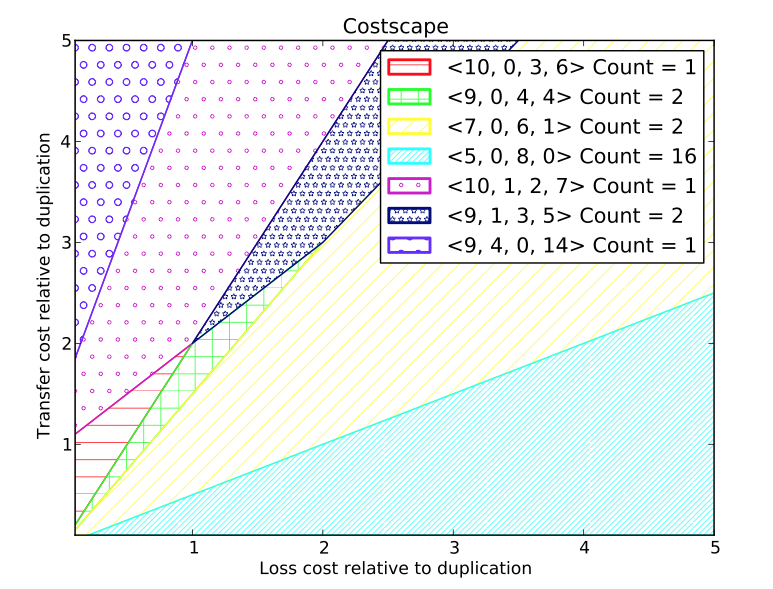
- costscape: This tool renders the cost space and partitions it into regions, where each region admits the same set of maximum parsimony reconciliations. The regions are listed (along with a count of the number of distinct reconciliations) and two-dimensional color-coded rendering is displayed or saved to a file. An example of such a rendering is shown at right.
- eventscape: This tool determines the individual events that are common to every reconciliation in each region. In addition, it collects all of those events and partitions them into those that are found in exaclty one region, exactly two regions, and up to all of the regions. This tool is useful in identifying the events that are highly supported by merit of occuring in multiple regions. The output is saved in a .csv file that can be opened and manipulated in Excel.
- sigscape: This tool uses
permutation testing to determine empirical p-values for significance
testing. While standard tests use a single set of event costs, this
tool computes p-values for entire event cost space. The tool
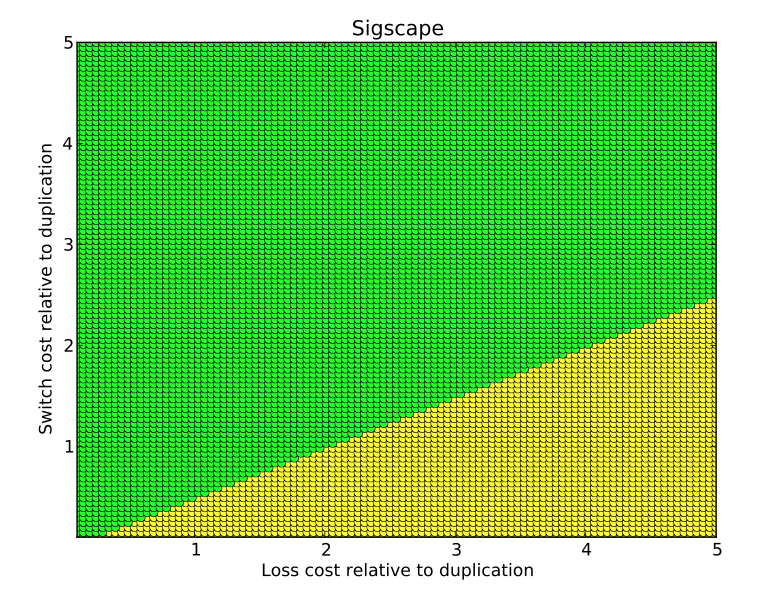 reports what fraction of the space achieves significance at the 0.01
level, between 0.01 and 0.05, and below the 0.05 level. A
color-coded plot is stored where green, yellow, and red represent
these three levels.
reports what fraction of the space achieves significance at the 0.01
level, between 0.01 and 0.05, and below the 0.05 level. A
color-coded plot is stored where green, yellow, and red represent
these three levels.
Installation, Requirements, and Usage
Xscape is available as a web-based application (no installation required) and as a downloadable set of Python tools.
1. Web interface for Xscape
This is the easiest way to use the tools. No downloading or installation is required. You'll be prompted to create an account which will allow you to upload your files. choose the appropriate tool and parameters, and your results will be sent back to you by e-mail.
2. Download Xscape
Downloading the software requires a bit of work installing the appropriate Python packages. Users who want to run the tools on their own computers or wish to modify the code to their needs may prefer this option.
Implementation Details
The aforementioned paper on which these tools are based describes a dynamic programming algorithm for computing the Pareto-optimal event count vectors and a second algorithm for computer their associated regions. The current implementations of these tools use a slightly different formulation based on the technical report CS-2011-1 "Faster Dynamic Programming Algorithms for the Cophylogeny Reconstruction Problem" by A. Yodpinyanee, B. Cousins, J. Peebles, T. Schramm, and R. Libeskind-Hadas.
The current release of the code uses memoization rather rather than dynamic programming. The region computation is done using a quadratic time polygon-intersection algorithm rather than the theoretically optimal O(N log N) divide-and-conquer algorithm. Finally, the current implementation of sigscape uses uniform sampling rather than the analytic solution.
Acknowledgements
The authors acknowledge the contributions of Dr. Matthew Rasmussen, who developed some of the software used in the tanglegram viewer. The first author gratefully acknowledges long-term collaborator Dr. Michael Charleston for many useful conversations and for his early work on Pareto-optimality in maximum parsimony reconciliation.
Questions, Comments, and Suggestions
Please contact Ran Libeskind-Hadas at (firstname AT cs DOT hmc DOT edu) with any questions, comments, or suggestions.